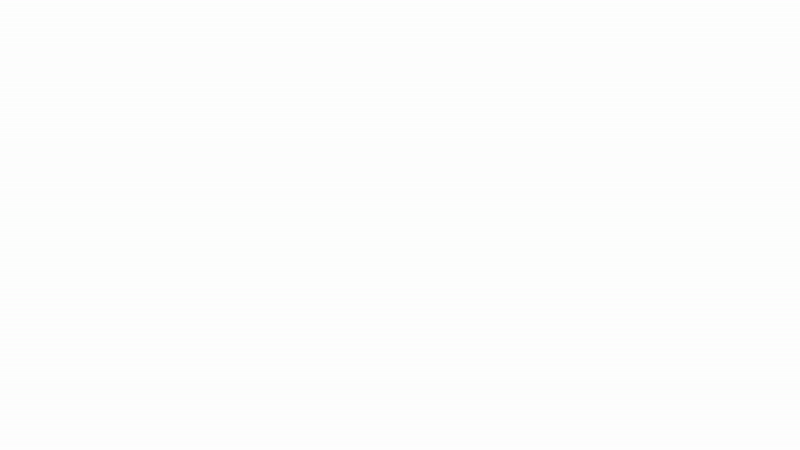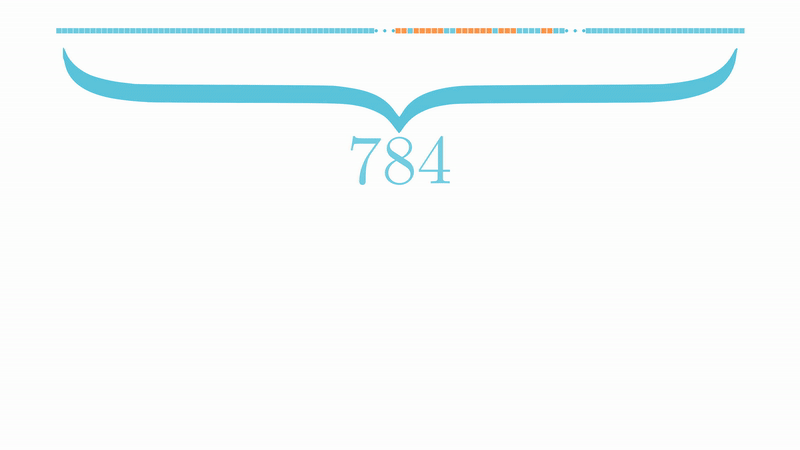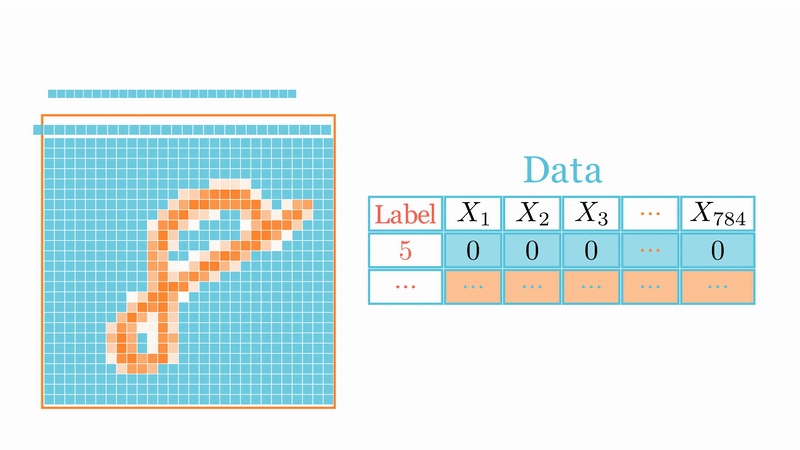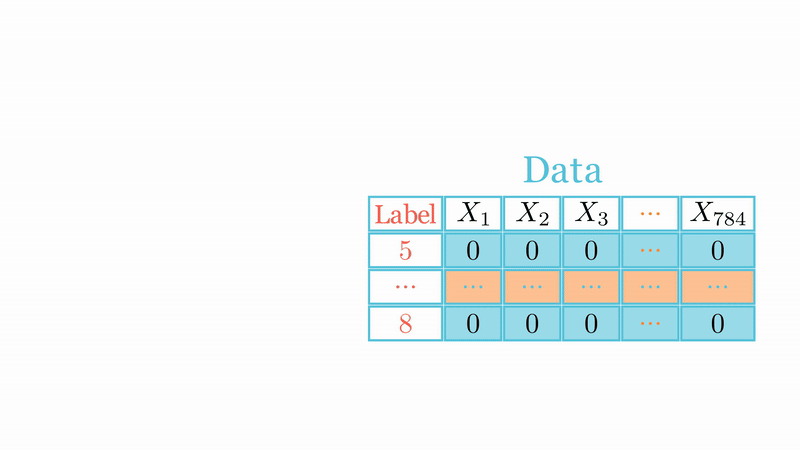Let \(X=\{\text{x}_1,\dots,\text{x}_n\}\subset\mathbb{R}^d\) be \(n\) points in \(d\)-dimensional Euclidean space \(\mathbb{R}^d\), and let \({\cal G}=(G_{i,j})\) with \(G_{k,j}\overset{iid}{\sim}{\cal N}(0,1/d')\) for \(k=1,\dots,d\) and \(j=1,\dots,\color{blue}{d'}\) with \(\color{blue}{d'}<d\). Let also \(\widehat{\text{x}}_i={\cal G}\text{x}_i\) and \(\widehat{\text{x}}_j={\cal G}\text{x}_j\) be two points of \(\mathbb{R}^{d'}\), thus for any \(\varepsilon\in (0,1)\) one has \[\mathbb{P}_{\cal G}\left(\left|\frac{\|\widehat{\text{x}}_i-\widehat{\text{x}}_j\|_{\color{blue}{d'}}^2}{\|\text{x}_i-\text{x}_j\|_d^2}-1,\forall i,j\right|> \varepsilon\right)< 2ne^{-\color{blue}{d'}(\varepsilon^2/2-\varepsilon^3/3)}.\]